GSOC 2021 Final Report
This blog describes the work I did in summer 2021 as part of Google Summer of Code. See the project proposal and the first post for the proposed work. In short, I contributed spatial simulation algorithms to DiffEqJump – a Julia module for simulating jump processes. Below is a rather detailed description of all the work I have done this summer. Follow the embedded links for more details.
Here is a cool-looking simulation I made in 2019 as part of my BA thesis. 
Interface for spatial SSAs
Since DiffEqJump did not have spatial solvers before this summer, a new interface had to be developed. It had to be consistent with the existing interface because it feeds into the rich ecosystem of SciML, and be flexible enough for more spatial SSAs to be added. After a lot of trial and error such an interface was settled on. In short, only two extra keyword arguments are required of the user over what is needed for an ordinary non-spatial problem. See the previous post or this tutorial for a detailed description of the interface.
Apart from the interface that is exposed to the user, an internal interface for spatial SSAs was developed. The most notable elements of it are three structs that keep track of spatial information needed by any spatial solver: CartesianGrid, RxRates and HopRates.
Spatial Solvers
Two specialized spatial solvers were added to DiffEqJump – NSM and DirectCRDirect. Additionally, a flattening method was added as fallback. See post 4 for a description of the two solvers and flattening. In short, NSM uses a binary heap to choose the next site to fire, resulting in asymptotic runtime – the time to update the heap; DirectCRDirect uses a clever priority table resulting in runtime but with higher overhead; flattening treats species at different sites as distinct and treats diffusive hops as monomolecular reactions resulting in extremely high memory usage. The asymptotic runtime of flattened algorithms depend on the exact non-spatial SSA used but are not representative of real performance due to inefficient memory use.
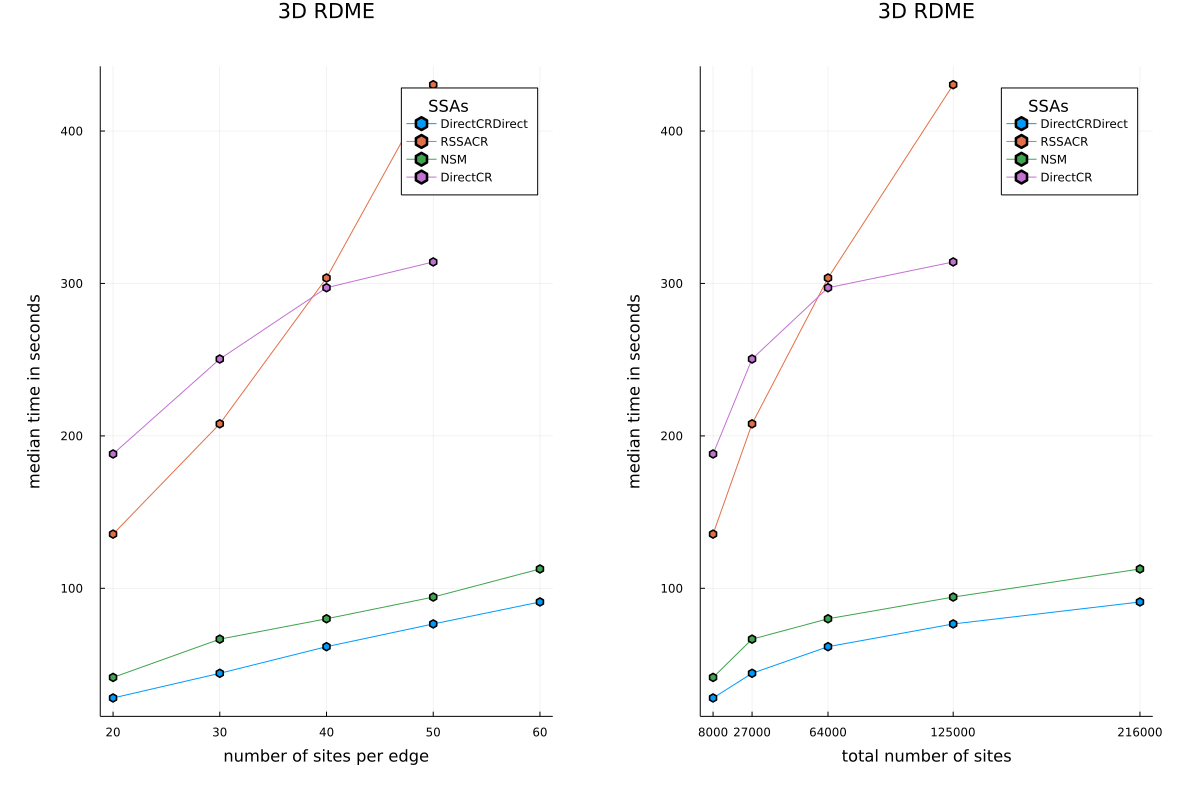
Both specialized solvers perform better than the two best existing non-spatial solvers – RSSACR and DirectCR as can be seen in the figure below.
Spatial structs and utilities
There are now four files with utility functions and structs for spatial solvers – utils.jl, topology.jl, hop_rates.jl and reaction_rates.jl.
The first one contains functions for doing various routines common to all or many spatial SSAs. For example update_state! updates the current state.
The second file contains several structs to keep track of the topology of the system. Most notably, CartesianGrid represents a Cartesian Grid – an interval, a rectangle, or a 3D equivalent of a rectangle (sometimes called hyperrectangle). See post 3 for a description of CartesianGrid. Additionally, any graph from LightGraphs can be passed in, allowing for simulations on unstructured meshes. This can be useful in epidemics simulations (representing a geographical region as a graph) and simulations on meshes obtained from triangulating a surface.
The last two files implement two analogous structs – RxRates to keep track of reactions and HopRates to keep track of spatial hops. The latter of the two is specifically optimized for CartesianGrid.
Tests, tutorials, benchmarks, documentation
In order to ensure accuracy of the newly added solvers and utility structures three tests have been created. One tests utilities and the other two test the accuracy of the solvers. All three tests are passing.
A comprehensive tutorial has been written and added to SciMLTutorials. This will help users get started with the new functionality.
A benchmarking script has been written, and will be included in SciMLBenchmarks. It uses a model from a research paper and compares the performance of the newly added spatial SSAs to the best existing SSAs. The figure above is generated by this script.
All functions and structures have docstrings in order to help future developers understand and extend the code.
Further work
This issue on Github contains the roadmap of what has been done and what is left to do in the future. Some of the most important items to be implemented in the future are:
escaping boundary condition
other types of jumps such as constant-rate jumps
spatially varying mass-action jumps
Tooling for generating hopping rates for diffusion, advection-diffusion, and drift-diffusion
spatially distributed reactions (e.g. CRDME)
List of all Pull Requests
Here is the complete list of my pull requests to DiffEqJump. All of them have been merged.
Next Subvolume Method on Graphs – implemented
NSM, wrote two tests, and established some of the basic interface, a lot of which was reworked later on.Spatial TODOs – added optimized versions of
CartesianGrid, wrote a better test, implemented a more general form of hopping rates.added
reset!forPriorityTable– small PR adding one function.DirectCRonDirect – implemented the second spatial SSA –
DirectCRDirect.Improved
HopRates– optimizedHopRates.Made
tstopstype-stable inSSAStepper– a small PR fixing a minor type-instability.Cosmetic improvements – a purely cosmetic PR: renaming things, adding docstrings, etc.
Improved
HopRates– optimizedHopRatesforCartesianGrid.More
HopRates– added two new forms of hopping rates to reduce memory use.Rename file – small PR renaming a file.
New
PriorityTable– rewrote thePriorityTablestructure to make it faster. This PR will be merged once it passes review.
I made two pull requests to other SciML repositories – SciMLTutorials and SciMLBenchmarks.
Tutorial for the spatial reversible binding jump model – a tutorial showing how to use the newly added functionality in
DiffEqJump. This was merged.Benchmarking – a benchmarking script comparing the spatial SSAs. It will be merged once it passes review.